|
|
|
| Home » Molecular Genetics » Research |
| Molecular Genetics |
| Sex chromosome evolution |
Sex chromosomes differ from autosomes in gene content, organization and expression owing to several reasons such as, absence of crossing over, dosage compensation, meiotic sex chromosome inactivation, hemizygous condition in one of the sexes, etc. Several studies in different model and non-model organisms have given evidence to this. The sheer diversity in sex chromosomes in different animal species makes it hard to find a common pattern for evolution of sex chromosomes. Sex chromosomes, apart from their role in sex determination, are also involved in manifestation of sexually dimorphic traits. They are evolving independently in different species from autosomes, yet they are found to follow a common evolutionary path and thus representing a fascinating example of convergent evolution.
Silkworm offers an unique system to study these evolutionary processes as it has contrasting sex chromosome composition when compared to other well-known model organisms such as mouse and fruit fly. We are interested in studying the architecture of sex chromosomes in this female heterogametic system
Silkworm Z chromosome is masculinized and defeminized
Through genomic and transcriptomic approaches we have shown that silkworm Z chromosome harbors significantly high number of male specific genes. Contrarily, mapping female biased genes onto chromosomes revealed that Z chromosome is almost devoid of female biased genes.
Faster-Z evolution in silkmoths
Theoretical work predicts natural selection to be more efficient in the fixation of beneficial mutations in X/Z -linked genes than in autosomal genes. This .fast-X/Z effect. should be evident by an increased ratio of non-synonymous to synonymous substitutions (dN/dS) for sex-linked genes; however, recent studies have produced mixed support for this expectation. To make an independent test of the idea of fast-X/Z evolution, we envisage studying silkworm Z chromosome genes in comparison with those of autosomes, where empirical evidence would predict Fast Z effect.
|
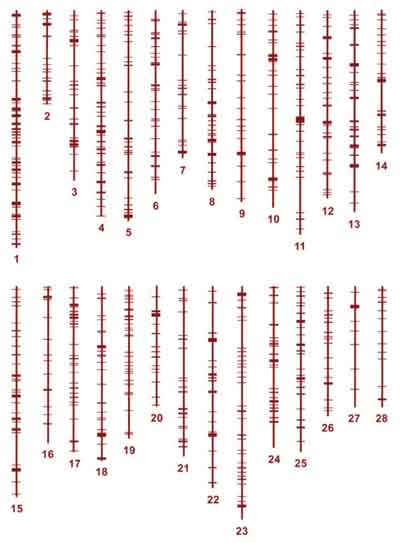 |
Physical map showing the distribution testis-specific genes on Bombyx mori chromosomes (Arunkumar et al., 2009, Genetics)
Further reading
Sackton TB, Corbett-Detig RB, Nagaraju J, Vaishna RL, Arunkumar KP and Hartl DL (2014) Positive selection drives faster-Z evolution in silkmoths. Evolution 68: 2331-2342
Suetsugu Y, Futahashi R, Kanamori H, . Arunkumar KP, Tomar A, Nagaraju J, Goldsmith MR, Feng Q, Xia Q, Yamamoto K, Shimada T and Mita K (2013) Large Scale Full-Length cDNA Sequencing Reveals a Unique Genomic Landscape in a Lepidopteran Model Insect, Bombyx mori. G3:Genes, Genomes, Genetics 3:1481-1492
Arunkumar KP, Mita K and Nagaraju J (2009) Silkworm testis specific genes are enriched on Z chromosome and are evolutionarily conserved. Genetics 182: 493-501.
|
|
| Last updated on: Friday, 21th Nov, 2014. |
|
|
|
|