|
|
|
| Home »
Genomics and Profiling Applications » Research |
| Genomics and Profiling Applications |
Projects
|
Human genetic variation studies in Indian populations.
India represents about one-sixth of the world population and comprise of people from diverse linguistic families, cultural practices and genetic architecture. Anthropological and genetic studies have shown that the current peopling of the Indian subcontinent has been characterized by a complex demographic history with contributions from different ancient human populations. The Indian populations have been hugely underrepresented in detailed genomic studies and hence, the genetic makeup of the diverse Indian populations is largely unknown. Studying the human genetic diversity in various Indian populations is crucial and have implications in several aspects of human genetics including medical genetics, phylogeography, population structure and DNA forensics.
The major research interests of our group include studying the genetic diversity among the Indian populations and utilize them for improving the current human DNA profiling technology for forensic human identification (HID). In this direction, our group is working on supplementing the ubiquitously employed short tandem repeats (STRs) with additional markers to enhance the success rate of DNA profiling while encountering challenging forensic samples (such as exhumed bones and burnt tissues). Recently, we designed a panel comprising of SNPs with desired features for HID application for Indian populations (Figure 1), with much better performance parameters than the currently employed STR-based panels and other SNP-panels proposed elsewhere. In addition to SNPs, we are also employing the applicability of additional autosomal and Y-chromosomal STRs (now available in the form of commercial kits).
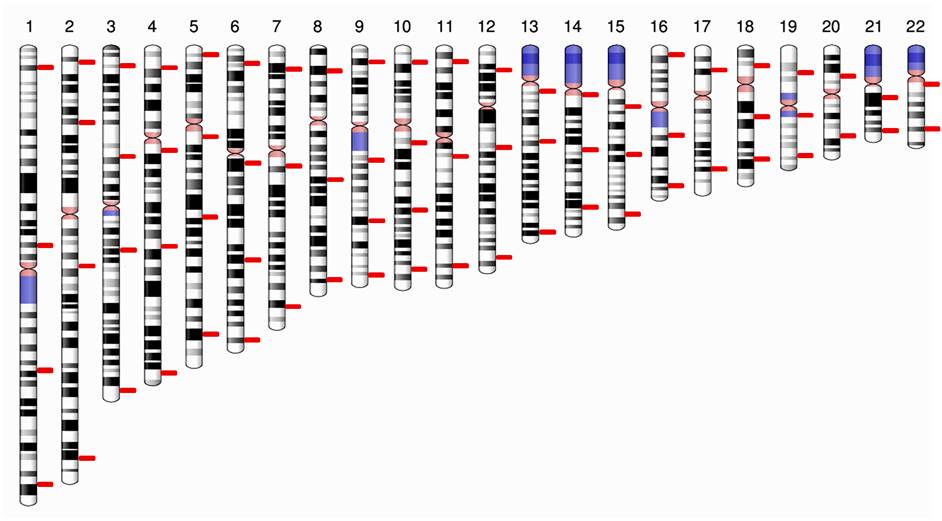
Figure 1. Diagrammatic representation of the 70 SNPs (shown with red bars) on the autosomes based upon their approximate locations (Sarkar and Nandineni, 2017)
Another aspect of our work focuses on genotype-phenotype correlation studies to validate the association of DNA-based markers (especially SNPs) with common externally visible characteristics (EVCs) like human skin colour, body-mass index (BMI) and height. Such studies will aid in understanding the association of SNPs and common phenotypes in Indian populations which were otherwise, highly underrepresented in previous studies. These studies have forensic implications as well since a better understanding of the effects of genetic variants could reliably predict the phenotype based on DNA evidence (often termed as ‘forensic phenotyping’). Such predictions will help the forensic community to generate an investigative lead in cases where there are no apparent suspects or database hits.
Apart from studying genetic variations based on human genomic DNA, we are also studying the human salivary microbiome diversity in various Indian populations by employing a high-throughput sequencing approach. Currently, by analyzing the variable regions of the phylogenetic marker 16S rRNA gene, we are identifying the occurrence and distribution of various bacterial genera in Indian populations. This will help to understand the salivary microbiome diversity in Indian populations when compared to the other worldwide populations, its association with physical and climatic factors and to address whether the Indian populations possess a unique ‘core microbiome’.
Plant-fungal interaction studies in the chilli-Colletotrichum pathosystem.
Chilli, a native of Central and South America, is an indispensable spice in everyday cuisine all over the world. It is rich in vitamin C and imparts a characteristic piquancy to the food due to the presence of capsaicin. Capsicum annuum L., family Solanaceae, is an economically important cultivated species of chilli and is the most widely grown spice in the world. India has the largest area under cultivation for chilli and is the world leader in its production, consumption and export, with states of Andhra Pradesh and Telangana contributing to ~49% of total production. One of the most devastating diseases and a major economic constraint to chilli production, especially in tropical and subtropical regions of the world, including India is anthracnose disease. It is caused by Colletotrichum truncatum (formerly called C. capsici), a fungal pathogen belonging to the Ascomycota. The disease is characterized by dark, sunken necrotic lesions with concentric rings of acervuli, containing curved conidiospores that form specialized structures for host penetration called appresoria; thus reducing the quality and marketability of chilli fruits (Figure 2).
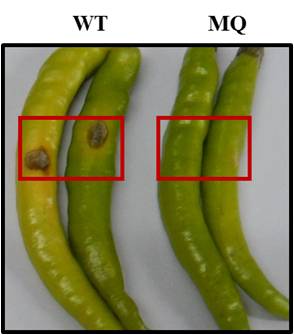
Figure 2. Pathogenicity assay of C. truncatum on chillies. The points of inoculation are shown in red boxes. The chillies inoculated with wild type (WT) fungal conidia show necrotic lesions. The chillies inoculated with MilliQ water (MQ) are taken as control.
The natural resistance to anthracnose is observed in only two species of chilli viz., C. chinense and C. baccatum. Traditional control measures like use of fungicides have not been very sustainable and no resistant cultivars of C. annuum have successfully been developed so far. The molecular mechanism of disease development in chilli upon infection with C. truncatum has not yet been elucidated in detail. Some recent studies suggest that this pathogen follows a subcuticular/intramural colonization strategy and has asymptomatic endophytic phase after initial infection and prior to necrotrophic development. However, it contrasts the intracellular hemibiotrophic infection strategy of most other Colletotrichum species, where the pathogen first establishes intracellular biotrophic hyphae inside living host cells before switching to destructive necrotrophy. With the availability of whole genome sequence for chilli and many Colletotrichum species, the chilli - C. truncatum pathosystem provides an excellent model for studies of the infection process and molecular interactions between the host and pathogen.
Our laboratory is interested in identifying and characterizing pathogenicity genes in C. truncatum to get an insight into different aspects of its infection biology, life-style and host specificity using two approaches: (i) random insertional mutagenesis for identification of loss-of-function mutants and (ii) whole genome/transcriptome sequencing of the C. truncatum genome. Agrobacterium tumefaciens-mediated transformation (ATMT) is employed to generate a library of random insertional mutants that are being screened for altered (partial or complete loss of) pathogenicity phenotype on chilli to identify the putative pathogenicity genes. The de novo whole genome sequence of C. truncatum has been obtained using the Illumina HiSeq platform with a combination of both short insert paired-end and long insert mate-pair libraries. The de novo transcriptome assembly of C. truncatum is being used for evidence-based gene calling in combination with different ab initio gene callers. Functional annotation of gene models is being carried out in order to identify some novel effectors and genes involved in the pathogenicity and virulence of C. truncatum that could be responsible for its host specific life-style and infection strategy. The putative pathogenicity genes identified through ATMT and NGS analysis would be validated by targeted gene knockout. The differential expression of the validated pathogenicity genes of C. truncatum would be studied during different stages of the anthracnose development on chilli. This study is expected to help in developing efficient control strategies for anthracnose disease on chilli and other related crops in future and suggest novel disease control measures based on chemical interventions or by breeding of anthracnose-resistant chillies.
|
|
|
| Contact Information |
| Email: |
|
| Phone: |
+91-40-27216141 (Direct), +91-40-27216142 (Secretary)
+91-40-27216143/144 (Lab)
|
| Fax: |
+91-40- 27216006 |
|
| Last updated on: Friday, 13th January, 2017. |
|
|
|
|