|
APEDA - CDFD Centre for Basmati DNA analysis
Agricultural and Processed Food Products Export Development Authority (APEDA) - Centre for DNA Fingerprinting & Diagnostics (CDFD) Centre for Basmati DNA analysis was established in CDFD in 2005 to test Basmati rice samples using the DNA protocol developed at CDFD. The initial funds for establishment of the laboratory have been provided by Basmati Export Development Foundation (BEDF) managed by APEDA, Ministry of Commerce, Government of India.
At the APEDA-CDFD centre, the Basmati rice samples, received from Export Inspection Council (EIC), Ministry of commerce, Govt. of India, Basmati rice exporters and Basmati importers from India and other countries, are tested for their purity by using the DNA protocol developed and standardized at CDFD. The Basmati DNA Test Report indicating the level of purity of Basmati rice in a given sample and the extent of adulteration with non-Basmati rice grains, if any, will be indicated
Service charges:
₹ 12,000/- (Rupees twelve thousands only) Plus 18% Service Tax (as per GST) that amounts to ₹ 14,160/- (Rupees fourteen thousand one hundred sixty only)
Management Committee of the APEDA-CDFD Centre for Basmati DNA Analysis
Director
Chairperson
(DBT, Ministry of Science & Technology, Govt. of India)
Hyderabad
Chairman
Co-Chairperson
Agricultural and Processed Food Products Export Development Authority (APEDA)
(Ministry of Commerce & Industry, Govt. of India)
New Delhi
Director
Member
Export Inspection Council of India (EIC)
(Ministry of Commerce & Industry, Govt. of India)
New Delhi
Executive Director
Member
All India Rice Exporters Association
New Delhi
Prof. E.A. Siddiq
Member
Distinguished Chair
Acharya N.G. Ranga Agricultural University and CDFD
Hyderabad
Director
Tilda Rice Land Pvt. Limited
Gurgaon
Dr. Kasbekar
Consultant Scientist & Member Convener
Haldane Chair
Laboratory of Neurospora Genetics
Centre for DNA Fingerprinting and Diagnostics (CDFD)
Hyderabad
Protocol
|
Sampling procedure and DNA extraction
From the samples received at the Centre, at least 50 g of grains from each sample is powdered and the remaining sample is stored for three months. During this period, if required, the sample is re-used for DNA analysis. From the powdered sample, eight sub-samples of 1 g each is drawn (Fig. 1), from which 100 mg of grain powder is collected for DNA extraction. DNA is isolated using Qiagen DNeasy plant mini kit and quantity and quality of the DNA (at 260/230 and 260/280 absorbance) are verified using UV-Vis spectrophotometer.
PCR amplification
The PCR is performed using the "Basmati VerifilerTM Kit" (Labindia) (Licensed by CDFD to Ms. Labindia under license transfer agreement) and the protocol is followed as per the details provided therein. (PDF Basmati Verifiler Kit)
Capillary electrophoresis (CE)
The fluorescently labeled PCR products are mixed with 0.3 µl of GeneScan-500 ROX (6-Carboxy-X-Rhodamine) size standard (Applied Biosystems, USA) and 10.2 µl of Hi-Di Formamide (Applied Biosystems, USA), electrophoresed by capillary electrophoresis on an ABI PRISM 3100 genetic analyzer (Applied Biosystems, USA) according to the manufacturer's instructions. Subsequently, fluorescent DNA fragments are resolved using the GeneScan version 3.7, and allele size and peak-area of the true peaks are determined by Genemapper version 3.7 (Applied Biosystems, USA).
Note: - In case of adulterated samples, each experiment is repeated thrice, starting from the DNA extraction, to confirm the extent of adulteration of Basmati rice with non-basmati rice grains.
Quantification of adulteration
The quantity of an amplified PCR product is represented by peak area (measured in relative fluorescent units, rfu). The adulteration is expressed as a percent fraction to that of pure basmati:
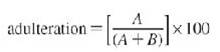
Where,
A is the rfu of the adulterant and
B is the rfu of the main variety.
Issue of Basmati DNA Test Report
Once the DNA analysis of a given rice sample is completed, the duly signed authenticity certificate, which contains the details of percentage of adulterated rice present in Basmati rice sample, is provided to the sender. The scanned copy of the certificate may also be sent by e-mail on request. The certificate is formatted as per the guidelines stipulated by the management committee of the APEDA-CDFD centre.
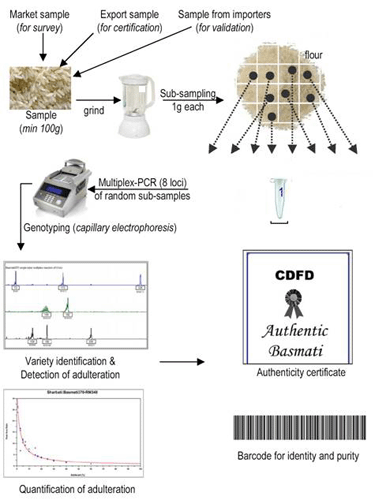
Fig. 1 Picture showing the protocol followed at the APEDA-CDFD centre for Basmati DNA analysis
Publications
IS 16538 : 2019, Microsatellite Based PCR Protocol for Identification of Basmati Rice Varieties, Bureau of Indian Standards, 2019
Lakshminarayana RV, Sabahat Noor, Satyavathi VV, Srividhya A, Kaliappan A, Parimala SRN, Bharathi PM, Deborah DA, Sudhakar Rao KV, Shobharani N, EA Siddiq EA, Javaregowda Nagaraju (2015) Discovery and mapping of genomic regions governing economically important traits of Basmati rice. BMC Plant Biology doi:10.1186/s12870-015-0575-5
Lakshminarayana RV, Satyavathi VV, EA Siddiq and Nagaraju J (2014) Review of methods for the detection and quantification of adulteration of rice: Basmati as a case study. Journal of Food Science and Technology DOI: 10.1007/s13197-014-1579-0
Archak S, Nagaraju J (2014) Computational analyses of protein coded by rice (Oryza sativa japonica) cDNA (GI: 32984786) indicate lectin like Ca(2+) binding properties for Eicosapenta Peptide Repeats (EPRs). Bioinformation 10:63-67.
Choudhary G, Ranjitkumar N, Surapaneni M, Deborah DA, Vipparla A, Anuradha G, Siddiq EA, Vemireddy LR (2013) Molecular genetic diversity of major Indian rice cultivars over decadal periods. PLoS One 8:e6619.
Siddiq EA, Vemireddy LR and Nagaraju J (2012) Basmati rices: genetics, breeding and trade. Agricultural Research 1: 25-36.
Sinha DK, Nagaraju J, Tomar A, Bentur JS and Nair S (2012) Pyrosequencing-based transcriptome analysis of the Asian Rice Gall Midge reveals differential response during compatible and incompatible interaction. International Journal of Molecular Science 13:13079-13103.
Archak S, Nagaraju J (2007) Computational prediction of rice (Oryza sativa) miRNA targets. Genomics, Proteomics and Bioinformatics 5: 196-206.
Lakshminarayanareddy V, Archak S, Nagaraju J (2007) Capillary electrophoresis is essential for microsatellite marker based detection and quantification of adulteration of Basmati rice (Oryza sativa). Journal of Agriculture and Food Chemistry 55: 8112-8117.
Archak S, Lakshminarayanareddy V, Nagaraju J (2007) High-throughput multiplex microsatellite marker assay for detection and quantification of adulteration in Basmati rice (Oryza sativa). Electrophoresis 28: 2396-2405.
Nagaraju J, Kathirvel M, Kumar RR, Siddiq EA, Hasnain SE (2002) Genetic analysis of traditional and evolved Basmati and non-Basmati rice varieties by using fluorescence-based ISSR-PCR and SSR markers. Proceedings of the National Academy of Sciences USA 99: 5836-5841.
Patents taken/applied
J. Nagaraju . An Indian patent has also been filed with PCT for DNA markers, which distinguish traditional Basmati, evolved Basmati and non-Basmati rice varieties. (Patent No 260/MAS/2002, dated April 8, 2002)
J. Nagaraju . The US patent has been applied for multiplex Simple Sequence Repeat kit containing 10 loci which can simultaneously distinguish traditional Basmati, evolved Basmati and non-Basmati varieties and quantify the adulterants (File No.FPO2559/RB/SM dated 19.04.2006)
J. Nagaraju. High-throughput multiplex microsatellite marker assay for detection and quantification of adulteration in Basmati rice (Indian Patent No.260/MAS/2002 and 662/CHE/2006; USA Patent No.USPTO 10/357, 488 and 11/406, 257; International Patent No.PCT/IN06/00254)
Related Publications
Masouleh AK, Waters DLE, Reinke RF and Henry RJ (2009) A high-throughput assay for rapid and simultaneous analysis of perfect markers for important quality and agronomic traits in rice using multiplexed MALDI-TOF mass spectrometry. Plant Biotechnology Journal 7:355-363
Aggarwal RK, Shenoy VV Ramadevi J, Rajkumar R and Singh L (2002) Molecular characterization of some Indian Basmati and other elite rice genotypes using fluorescent-AFLP.Theor Appl Genet 105:680-690.
Bligh HFJ (2000) Detection of adulteration of Basmati rice with non-premium long-grain rice. International Journal of Food Science & Technology 35: 257-265.
Choudhury PR, Kohli S, Srinivasan K, Mohapatra T and Sharma RP (2001). Identification and classification of aromatic rices based on DNA Fingerprinting. Euphytica 118:243-251.
Colyer A, Macarthur R, Lloyd J and Hird H (2008). Comparison of calibration methods for the quantification of Basmati and non-Basmati rice using microsatellite analysis. Food Additives and Contaminants 1-6.
Garris AJ, Tai TH, Coburn J, Kresovich S and McCouch S (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169: 1631.1638.
Glaszmann JC (1987) Isozymes and classification of Asian rice varieties. TAG Theoretical and Applied Genetics 74:21-30.
Jain S, Jain RK and McCouch SR (2004) Genetic analysis of Indian aromatic and quality rice (Oryza sativa L.) germplasm using panels of fluorescently-labelled microsatellite markers. Theor Appl Genet 109: 965-977.
Kelly S, Baxter M, Chapman S, Rhodes C, Dennis J and Brereton P (2002) The application of isotopic and elemental analysis to determine the geographical origin of premium long grain rice. Eur Food Res Technol 214:72-78.
Kelly S, Heaton K and Hoogewerff J (2005) Tracing the geographical origin of food: The application of multi-element and multi-isotope analysis Trends in Food Science & Technology 16:555-567.
Lapitan VC, Brar DS, Abe T and Redona ED (2007). Assessment of genetic diversity of Philippine rice cultivars carrying good quality traits using SSR markers. Breeding Science 57:263-270.
Lopez SJ (2008). TaqMan based real time PCR method for quantitative detection of basmati rice adulteration with non-basmati rice. Eur Food Res Technol 227:619-622.
Siwach P, Jain S, Saini N, Chowdhury VK and Jain RK (2004). Allelic diversity among Basmati and Non-Basmati long-grain indica rice varieties using microsatellite markers. J. Plant Biochemistry & Biotechnology 13:25-32.
Steele KA, Ogden R, McEwing R, Briggs H, and Gorham J (2008) InDel markers distinguish Basmatis from other fragrant rice varieties. Field Crops Research 105:81-87.
Woolfe M and Primrose S (2004) Food forensics: using DNA technology to combat misdescription and fraud. TRENDS in Biotechnology 22(5):222-226.
Quality Standardization of Indian Basmati Rice Exports. (1999) ASIA-PACIFIC BIOTECH NEWS (APBN) 3(18):431.
|
|